Eaton & Ree (2013) single-end RAD data set
Here we demonstrate a denovo assembly for an empirical RAD data set using the ipyrad Python API. This example was run on a workstation with 20 cores and takes about 10 minutes to assemble, but you should be able to run it on a 4-core laptop in ~30-60 minutes.
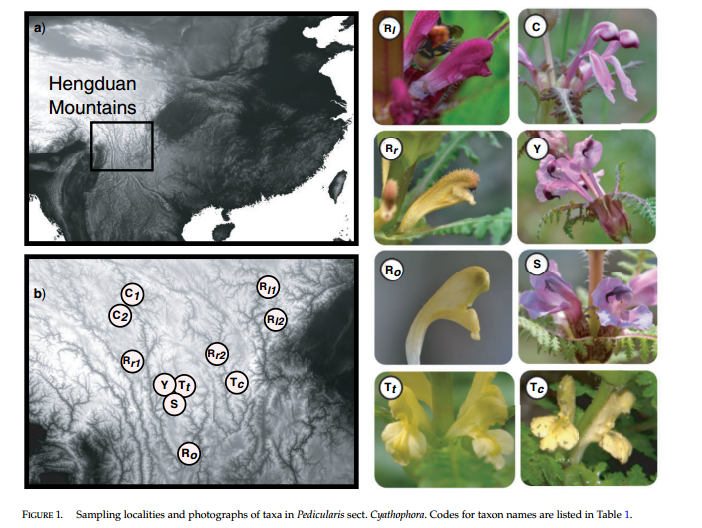
For our example data we will use the 13 taxa Pedicularis data set from Eaton and Ree (2013) (Open Access). This data set is composed of single-end 75bp reads from a RAD-seq library prepared with the PstI enzyme. The data set also serves as an example for several of our analysis cookbooks that demonstrate methods for analyzing RAD-seq data files. At the end of this notebook there are also several examples of how to use the ipyrad analysis tools to run downstream analyses in parallel.
Setup (software and data files)
If you haven’t done so yet, start by installing ipyrad using conda (see ipyrad installation instructions) as well as the packages in the cell below. This is easiest to do in a terminal. Then open a jupyter-notebook, like this one, and follow along with the tuturial by copying and executing the code in the cells, and adding your own documentation between them using markdown. Feel free to modify parameters to see their effects on the downstream results.
[4]:
## conda install ipyrad -c ipyrad
## conda install toytree -c eaton-lab
## conda install sra-tools -c bioconda
[5]:
## imports
import ipyrad as ip
import ipyrad.analysis as ipa
import ipyparallel as ipp
In contrast to the ipyrad CLI, the ipyrad API gives users much more fine-scale control over the parallelization of their analysis, but this also requires learning a little bit about the library that we use to do this, called ipyparallel. This library is designed for use with jupyter-notebooks to allow massive-scale multi-processing while working interactively.
Understanding the nuts and bolts of it might take a little while, but it is fairly easy to get started using it, especially in the way it is integrated with ipyrad. To start a parallel client to you must run the command-line program ‘ipcluster’. This will essentially start a number of independent Python processes (kernels) which we can then send bits of work to do. The cluster can be stopped and restarted independently of this notebook, which is convenient for working on a cluster where
connecting to many cores is not always immediately available.
[6]:
# Open a terminal and type the following command to start
# an ipcluster instance with 40 engines:
# ipcluster start -n 40 --cluster-id="ipyrad" --daemonize
# After the cluster is running you can attach to it with ipyparallel
ipyclient = ipp.Client(cluster_id="ipyrad")
Download the data set (Pedicularis)
These data are archived on the NCBI sequence read archive (SRA) under accession id SRP021469. As part of the ipyrad analysis tools we have a wrapper around the SRAtools software that can be used to query NCBI and download sequence data based on accession IDs. Run the code below to download the fastq data files associated with this study. The data will be saved the specified directory which will be created if it does not already exist. The compressed file size of the data is a little over 1GB. If
you pass your ipyclient to the .run() command below then the download will be parallelized.
[ ]:
## download the Pedicularis data set from NCBI
sra = ipa.sratools(accessions="SRP021469", workdir="fastqs-Ped")
sra.run(force=True, ipyclient=ipyclient)
[####################] 100% Downloading fastq files | 0:01:19 |
13 fastq files downloaded to /home/deren/Documents/ipyrad/tests/fastqs-Ped
Create an Assembly object
This object stores the parameters of the assembly and the organization of data files.
[6]:
## you must provide a name for the Assembly
data = ip.Assembly("pedicularis")
New Assembly: pedicularis
Set parameters for the Assembly. This will raise an error if any of the parameters are not allowed because they are the wrong type, or out of the allowed range.
[10]:
## set parameters
data.set_params("project_dir", "analysis-ipyrad")
data.set_params("sorted_fastq_path", "fastqs-Ped/*.fastq.gz")
data.set_params("clust_threshold", "0.90")
data.set_params("filter_adapters", "2")
data.set_params("max_Hs_consens", (5, 5))
data.set_params("trim_loci", (0, 5, 0, 0))
data.set_params("output_formats", "psvnkua")
## see/print all parameters
data.get_params()
0 assembly_name pedicularis
1 project_dir ./analysis-ipyrad
2 raw_fastq_path
3 barcodes_path
4 sorted_fastq_path ./example_empirical_rad/*.fastq.gz
5 assembly_method denovo
6 reference_sequence
7 datatype rad
8 restriction_overhang ('TGCAG', '')
9 max_low_qual_bases 5
10 phred_Qscore_offset 33
11 mindepth_statistical 6
12 mindepth_majrule 6
13 maxdepth 10000
14 clust_threshold 0.9
15 max_barcode_mismatch 0
16 filter_adapters 2
17 filter_min_trim_len 35
18 max_alleles_consens 2
19 max_Ns_consens (5, 5)
20 max_Hs_consens (5, 5)
21 min_samples_locus 4
22 max_SNPs_locus (20, 20)
23 max_Indels_locus (8, 8)
24 max_shared_Hs_locus 0.5
25 trim_reads (0, 0, 0, 0)
26 trim_loci (0, 5, 0, 0)
27 output_formats ('p', 's', 'v', 'n', 'k', 'u')
28 pop_assign_file
Assemble the data set
[24]:
## run steps 1 & 2 of the assembly
data.run("12")
Assembly: pedicularis
[####################] 100% loading reads | 0:00:04 | s1 |
[####################] 100% processing reads | 0:01:14 | s2 |
[25]:
## access the stats of the assembly (so far) from the .stats attribute
data.stats
[25]:
| state | reads_raw | reads_passed_filter | |
|---|---|---|---|
| 29154_superba | 2 | 696994 | 689996 |
| 30556_thamno | 2 | 1452316 | 1440314 |
| 30686_cyathophylla | 2 | 1253109 | 1206947 |
| 32082_przewalskii | 2 | 964244 | 955480 |
| 33413_thamno | 2 | 636625 | 626084 |
| 33588_przewalskii | 2 | 1002923 | 993873 |
| 35236_rex | 2 | 1803858 | 1787366 |
| 35855_rex | 2 | 1409843 | 1397068 |
| 38362_rex | 2 | 1391175 | 1379626 |
| 39618_rex | 2 | 822263 | 813990 |
| 40578_rex | 2 | 1707942 | 1695523 |
| 41478_cyathophylloides | 2 | 2199740 | 2185364 |
| 41954_cyathophylloides | 2 | 2199613 | 2176210 |
[26]:
## run steps 3-6 of the assembly
data.run("3456")
Assembly: pedicularis
[####################] 100% dereplicating | 0:00:07 | s3 |
[####################] 100% clustering | 0:05:08 | s3 |
[####################] 100% building clusters | 0:00:26 | s3 |
[####################] 100% chunking | 0:00:05 | s3 |
[####################] 100% aligning | 0:03:06 | s3 |
[####################] 100% concatenating | 0:00:15 | s3 |
[####################] 100% inferring [H, E] | 0:01:10 | s4 |
[####################] 100% calculating depths | 0:00:06 | s5 |
[####################] 100% chunking clusters | 0:00:06 | s5 |
[####################] 100% consens calling | 0:01:56 | s5 |
[####################] 100% concat/shuffle input | 0:00:05 | s6 |
[####################] 100% clustering across | 0:03:47 | s6 |
[####################] 100% building clusters | 0:00:06 | s6 |
[####################] 100% aligning clusters | 0:00:29 | s6 |
[####################] 100% database indels | 0:00:14 | s6 |
[####################] 100% indexing clusters | 0:00:03 | s6 |
[####################] 100% building database | 0:00:29 | s6 |
Branch to create several final data sets with different parameter settings
[11]:
## create a branch for outputs with min_samples = 4 (lots of missing data)
min4 = data.branch("min4")
min4.set_params("min_samples_locus", 4)
min4.run("7")
## create a branch for outputs with min_samples = 13 (no missing data)
min13 = data.branch("min13")
min13.set_params("min_samples_locus", 13)
min13.run("7")
## create a branch with no missing data for ingroups, but allow
## missing data in the outgroups by setting population assignments.
## The population min-sample values overrule the min-samples-locus param
pops = data.branch("min11-pops")
pops.populations = {
"ingroup": (11, [i for i in pops.samples if "prz" not in i]),
"outgroup" : (0, [i for i in pops.samples if "prz" in i]),
}
pops.run("7")
## create a branch with no missing data and with outgroups removed
nouts = data.branch("nouts_min11", subsamples=[i for i in pops.samples if "prz" not in i])
nouts.set_params("min_samples_locus", 11)
nouts.run("7")
Assembly: min4
[####################] 100% filtering loci | 0:00:08 | s7 |
[####################] 100% building loci/stats | 0:00:01 | s7 |
[####################] 100% building vcf file | 0:00:09 | s7 |
[####################] 100% writing vcf file | 0:00:00 | s7 |
[####################] 100% building arrays | 0:00:04 | s7 |
[####################] 100% writing outfiles | 0:00:05 | s7 |
Outfiles written to: ~/Documents/ipyrad/tests/analysis-ipyrad/min4_outfiles
Assembly: min13
[####################] 100% filtering loci | 0:00:02 | s7 |
[####################] 100% building loci/stats | 0:00:01 | s7 |
[####################] 100% building vcf file | 0:00:03 | s7 |
[####################] 100% writing vcf file | 0:00:00 | s7 |
[####################] 100% building arrays | 0:00:04 | s7 |
[####################] 100% writing outfiles | 0:00:00 | s7 |
Outfiles written to: ~/Documents/ipyrad/tests/analysis-ipyrad/min13_outfiles
Assembly: min11-pops
[####################] 100% filtering loci | 0:00:02 | s7 |
[####################] 100% building loci/stats | 0:00:01 | s7 |
[####################] 100% building vcf file | 0:00:03 | s7 |
[####################] 100% writing vcf file | 0:00:00 | s7 |
[####################] 100% building arrays | 0:00:04 | s7 |
[####################] 100% writing outfiles | 0:00:00 | s7 |
Outfiles written to: ~/Documents/ipyrad/tests/analysis-ipyrad/min11-pops_outfiles
Assembly: nouts_min11
[####################] 100% filtering loci | 0:00:03 | s7 |
[####################] 100% building loci/stats | 0:00:02 | s7 |
[####################] 100% building vcf file | 0:00:02 | s7 |
[####################] 100% writing vcf file | 0:00:00 | s7 |
[####################] 100% building arrays | 0:00:04 | s7 |
[####################] 100% writing outfiles | 0:00:00 | s7 |
Outfiles written to: ~/Documents/ipyrad/tests/analysis-ipyrad/nouts_min11_outfiles
View final stats
The .stats attribute shows a stats summary for each sample, and a number of stats dataframes can be accessed for each step from the .stats_dfs attribute of the Assembly.
[9]:
## we can access the stats summary as a pandas dataframes.
min4.stats
[9]:
| state | reads_raw | reads_passed_filter | clusters_total | clusters_hidepth | hetero_est | error_est | reads_consens | |
|---|---|---|---|---|---|---|---|---|
| 29154_superba | 6 | 696994 | 689996 | 134897 | 35143 | 0.010 | 0.002 | 34628 |
| 30556_thamno | 6 | 1452316 | 1440314 | 212960 | 52776 | 0.011 | 0.003 | 51701 |
| 30686_cyathophylla | 6 | 1253109 | 1206947 | 240824 | 54282 | 0.010 | 0.002 | 53347 |
| 32082_przewalskii | 6 | 964244 | 955480 | 151762 | 42487 | 0.013 | 0.002 | 41841 |
| 33413_thamno | 6 | 636625 | 626084 | 172709 | 31072 | 0.012 | 0.002 | 30674 |
| 33588_przewalskii | 6 | 1002923 | 993873 | 158933 | 46555 | 0.013 | 0.002 | 45892 |
| 35236_rex | 6 | 1803858 | 1787366 | 417632 | 54709 | 0.010 | 0.001 | 54064 |
| 35855_rex | 6 | 1409843 | 1397068 | 184661 | 56595 | 0.013 | 0.003 | 55371 |
| 38362_rex | 6 | 1391175 | 1379626 | 133541 | 53156 | 0.008 | 0.002 | 52496 |
| 39618_rex | 6 | 822263 | 813990 | 148107 | 44016 | 0.010 | 0.002 | 43409 |
| 40578_rex | 6 | 1707942 | 1695523 | 222328 | 56653 | 0.011 | 0.002 | 55937 |
| 41478_cyathophylloides | 6 | 2199740 | 2185364 | 173188 | 55208 | 0.008 | 0.001 | 54482 |
| 41954_cyathophylloides | 6 | 2199613 | 2176210 | 301343 | 76537 | 0.008 | 0.003 | 75167 |
[25]:
## or print the full stats file
cat $min4.stats_files.s7
## The number of loci caught by each filter.
## ipyrad API location: [assembly].stats_dfs.s7_filters
total_filters applied_order retained_loci
total_prefiltered_loci 96562 0 96562
filtered_by_rm_duplicates 2939 2939 93623
filtered_by_max_indels 189 189 93434
filtered_by_max_snps 679 18 93416
filtered_by_max_shared_het 865 708 92708
filtered_by_min_sample 47958 46646 46062
filtered_by_max_alleles 10694 4565 41497
total_filtered_loci 41497 0 41497
## The number of loci recovered for each Sample.
## ipyrad API location: [assembly].stats_dfs.s7_samples
sample_coverage
29154_superba 21054
30556_thamno 31790
30686_cyathophylla 26648
32082_przewalskii 13201
33413_thamno 18747
33588_przewalskii 15461
35236_rex 33228
35855_rex 33403
38362_rex 33748
39618_rex 28135
40578_rex 34107
41478_cyathophylloides 30826
41954_cyathophylloides 28260
## The number of loci for which N taxa have data.
## ipyrad API location: [assembly].stats_dfs.s7_loci
locus_coverage sum_coverage
1 0 0
2 0 0
3 0 0
4 5764 5764
5 4002 9766
6 3650 13416
7 3139 16555
8 3220 19775
9 4153 23928
10 4904 28832
11 5321 34153
12 4511 38664
13 2833 41497
## The distribution of SNPs (var and pis) per locus.
## var = Number of loci with n variable sites (pis + autapomorphies)
## pis = Number of loci with n parsimony informative site (minor allele in >1 sample)
## ipyrad API location: [assembly].stats_dfs.s7_snps
var sum_var pis sum_pis
0 2348 0 11768 0
1 4482 4482 10822 10822
2 5870 16222 7699 26220
3 6314 35164 4914 40962
4 5708 57996 2967 52830
5 4857 82281 1679 61225
6 3885 105591 874 66469
7 2932 126115 478 69815
8 1955 141755 188 71319
9 1255 153050 72 71967
10 827 161320 19 72157
11 506 166886 10 72267
12 262 170030 4 72315
13 141 171863 3 72354
14 78 172955 0 72354
15 39 173540 0 72354
16 19 173844 0 72354
17 10 174014 0 72354
18 6 174122 0 72354
19 2 174160 0 72354
20 1 174180 0 72354
## Final Sample stats summary
state reads_raw reads_passed_filter clusters_total clusters_hidepth hetero_est error_est reads_consens loci_in_assembly
29154_superba 7 696994 689996 134897 35143 0.010 0.002 34628 21054
30556_thamno 7 1452316 1440314 212960 52776 0.011 0.003 51701 31790
30686_cyathophylla 7 1253109 1206947 240824 54282 0.010 0.002 53347 26648
32082_przewalskii 7 964244 955480 151762 42487 0.013 0.002 41841 13201
33413_thamno 7 636625 626084 172709 31072 0.012 0.002 30674 18747
33588_przewalskii 7 1002923 993873 158933 46555 0.013 0.002 45892 15461
35236_rex 7 1803858 1787366 417632 54709 0.010 0.001 54064 33228
35855_rex 7 1409843 1397068 184661 56595 0.013 0.003 55371 33403
38362_rex 7 1391175 1379626 133541 53156 0.008 0.002 52496 33748
39618_rex 7 822263 813990 148107 44016 0.010 0.002 43409 28135
40578_rex 7 1707942 1695523 222328 56653 0.011 0.002 55937 34107
41478_cyathophylloides 7 2199740 2185364 173188 55208 0.008 0.001 54482 30826
41954_cyathophylloides 7 2199613 2176210 301343 76537 0.008 0.003 75167 28260
[13]:
## and we can access parts of the full stats outputs as dataframes
min4.stats_dfs.s7_samples
[13]:
| sample_coverage | |
|---|---|
| 29154_superba | 21054 |
| 30556_thamno | 31790 |
| 30686_cyathophylla | 26648 |
| 32082_przewalskii | 13201 |
| 33413_thamno | 18747 |
| 33588_przewalskii | 15461 |
| 35236_rex | 33228 |
| 35855_rex | 33403 |
| 38362_rex | 33748 |
| 39618_rex | 28135 |
| 40578_rex | 34107 |
| 41478_cyathophylloides | 30826 |
| 41954_cyathophylloides | 28260 |
[14]:
## compare this to the one above, coverage is more equal
min13.stats_dfs.s7_samples
[14]:
| sample_coverage | |
|---|---|
| 29154_superba | 2833 |
| 30556_thamno | 2833 |
| 30686_cyathophylla | 2833 |
| 32082_przewalskii | 2833 |
| 33413_thamno | 2833 |
| 33588_przewalskii | 2833 |
| 35236_rex | 2833 |
| 35855_rex | 2833 |
| 38362_rex | 2833 |
| 39618_rex | 2833 |
| 40578_rex | 2833 |
| 41478_cyathophylloides | 2833 |
| 41954_cyathophylloides | 2833 |
[15]:
## similarly, coverage is equal here among ingroups, but allows missing in outgroups
pops.stats_dfs.s7_samples
[15]:
| sample_coverage | |
|---|---|
| 29154_superba | 5796 |
| 30556_thamno | 5796 |
| 30686_cyathophylla | 5796 |
| 32082_przewalskii | 3266 |
| 33413_thamno | 5796 |
| 33588_przewalskii | 3747 |
| 35236_rex | 5796 |
| 35855_rex | 5796 |
| 38362_rex | 5796 |
| 39618_rex | 5796 |
| 40578_rex | 5796 |
| 41478_cyathophylloides | 5796 |
| 41954_cyathophylloides | 5796 |
Analysis tools
We have a lot more information about analysis tools in the ipyrad documentation. But here I’ll show just a quick example of how you can easily access the data files for these assemblies and use them in downstream analysis software. The ipyrad analysis tools include convenient wrappers to make it easier to parallelize analyses of RAD-seq data. Please see the full documentation for the ipyrad.analysis tools in the ipyrad documentation for more details.
[6]:
import ipyrad as ip
import ipyrad.analysis as ipa
[7]:
## you can re-load assemblies at a later time from their JSON file
min4 = ip.load_json("analysis-ipyrad/min4.json")
min13 = ip.load_json("analysis-ipyrad/min13.json")
nouts = ip.load_json("analysis-ipyrad/nouts_min11.json")
loading Assembly: min4
from saved path: ~/Documents/ipyrad/tests/analysis-ipyrad/min4.json
loading Assembly: min13
from saved path: ~/Documents/ipyrad/tests/analysis-ipyrad/min13.json
loading Assembly: nouts_min11
from saved path: ~/Documents/ipyrad/tests/analysis-ipyrad/nouts_min11.json
RAxML – ML concatenation tree inference
[17]:
## conda install raxml -c bioconda
## conda install toytree -c eaton-lab
[12]:
## create a raxml analysis object for the min13 data sets
rax = ipa.raxml(
name=min13.name,
data=min13.outfiles.phy,
workdir="analysis-raxml",
T=20,
N=100,
o=[i for i in min13.samples if "prz" in i],
)
[19]:
## print the raxml command and call it
print rax.command
rax.run(force=True)
raxmlHPC-PTHREADS-SSE3 -f a -T 20 -m GTRGAMMA -N 100 -x 12345 -p 54321 -n min13 -w /home/deren/Documents/ipyrad/tests/analysis-raxml -s /home/deren/Documents/ipyrad/tests/analysis-ipyrad/min13_outfiles/min13.phy -o 33588_przewalskii,32082_przewalskii
job min13 finished successfully
[13]:
## access the resulting tree files
rax.trees
[13]:
bestTree ~/Documents/ipyrad/tests/analysis-raxml/RAxML_bestTree.min13
bipartitions ~/Documents/ipyrad/tests/analysis-raxml/RAxML_bipartitions.min13
bipartitionsBranchLabels ~/Documents/ipyrad/tests/analysis-raxml/RAxML_bipartitionsBranchLabels.min13
bootstrap ~/Documents/ipyrad/tests/analysis-raxml/RAxML_bootstrap.min13
info ~/Documents/ipyrad/tests/analysis-raxml/RAxML_info.min13
[15]:
## plot a tree in the notebook with toytree
import toytree
tre = toytree.tree(rax.trees.bipartitions)
tre.draw(
width=350,
height=400,
node_labels=tre.get_node_values("support"),
);
tetrad – quartet tree inference
[16]:
## create a tetrad analysis object
tet = ipa.tetrad(
name=min4.name,
seqfile=min4.outfiles.snpsphy,
mapfile=min4.outfiles.snpsmap,
nboots=100,
)
loading seq array [13 taxa x 174180 bp]
max unlinked SNPs per quartet (nloci): 39149
[18]:
## run tree inference
tet.run(ipyclient)
host compute node: [20 cores] on tinus
inferring 715 induced quartet trees
[####################] 100% initial tree | 0:00:05 |
[####################] 100% boot 100 | 0:02:23 |
[19]:
## access tree files
tet.trees
[19]:
boots ~/Documents/ipyrad/tests/analysis-tetrad/min4.boots
cons ~/Documents/ipyrad/tests/analysis-tetrad/min4.cons
nhx ~/Documents/ipyrad/tests/analysis-tetrad/min4.nhx
tree ~/Documents/ipyrad/tests/analysis-tetrad/min4.tree
[22]:
## plot results (just like above, but unrooted by default)
## the consensus tree here differs from the ML tree above.
import toytree
qtre = toytree.tree(tet.trees.nhx)
qtre.root(wildcard="prz")
qtre.draw(
width=350,
height=400,
node_labels=qtre.get_node_values("support"),
);
STRUCTURE – population cluster inference
[ ]:
## conda install structure clumpp -c ipyrad
[25]:
## create a structure analysis object for the no-outgroup data set
## NB: As of v0.9.64 you can use instead: data=data.outfiles.snps_database
struct = ipa.structure(
name=nouts.name,
data="analysis-ipyrad/nouts_min11_outfiles/nouts_min11.snps.hdf5",
)
## set params for analysis (should be longer in real analyses)
struct.mainparams.burnin=1000
struct.mainparams.numreps=8000
[96]:
## run structure across 10 random replicates of sampled unlinked SNPs
for kpop in [2, 3, 4, 5, 6]:
struct.run(kpop=kpop, nreps=10, ipyclient=ipyclient)
submitted 10 structure jobs [nouts_min11-K-2]
submitted 10 structure jobs [nouts_min11-K-3]
submitted 10 structure jobs [nouts_min11-K-4]
submitted 10 structure jobs [nouts_min11-K-5]
submitted 10 structure jobs [nouts_min11-K-6]
[115]:
## wait for all of these jobs to finish
ipyclient.wait()
[115]:
True
[26]:
## collect results
tables = {}
for kpop in [2, 3, 4, 5, 6]:
tables[kpop] = struct.get_clumpp_table(kpop)
mean scores across 10 replicates.
mean scores across 10 replicates.
mean scores across 10 replicates.
mean scores across 10 replicates.
mean scores across 10 replicates.
[27]:
## custom sorting order
myorder = [
"41478_cyathophylloides",
"41954_cyathophylloides",
"29154_superba",
"30686_cyathophylla",
"33413_thamno",
"30556_thamno",
"35236_rex",
"40578_rex",
"35855_rex",
"39618_rex",
"38362_rex",
]
[33]:
## import toyplot (packaged with toytree)
import toyplot
## plot bars for each K-value (mean of 10 reps)
for kpop in [2, 3, 4, 5, 6]:
table = tables[kpop]
table = table.ix[myorder]
## plot barplot w/ hover
canvas, axes, mark = toyplot.bars(
table,
title=[[i] for i in table.index.tolist()],
width=400,
height=200,
yshow=False,
style={"stroke": toyplot.color.near_black},
)
TREEMIX – ML tree & admixture co-inference
[15]:
## conda install treemix -c ipyrad
[39]:
## group taxa into 'populations'
imap = {
"prz": ["32082_przewalskii", "33588_przewalskii"],
"cys": ["41478_cyathophylloides", "41954_cyathophylloides"],
"cya": ["30686_cyathophylla"],
"sup": ["29154_superba"],
"cup": ["33413_thamno"],
"tha": ["30556_thamno"],
"rck": ["35236_rex"],
"rex": ["35855_rex", "40578_rex"],
"lip": ["39618_rex", "38362_rex"],
}
## optional: loci will be filtered if they do not have data for at
## least N samples in each species. Minimums cannot be <1.
minmap = {
"prz": 2,
"cys": 2,
"cya": 1,
"sup": 1,
"cup": 1,
"tha": 1,
"rck": 1,
"rex": 2,
"lip": 2,
}
## sets a random number seed
import numpy
numpy.random.seed(12349876)
## create a treemix analysis object
tmix = ipa.treemix(
name=min13.name,
data=min13.outfiles.snpsphy,
mapfile=min13.outfiles.snpsmap,
imap=imap,
minmap=minmap,
)
## you can set additional parameter args here
tmix.params.root = "prz"
tmix.params.global_ = 1
## print the full params
tmix.params
[39]:
binary treemix
bootstrap 0
climb 0
cormig 0
g (None, None)
global_ 1
k 0
m 0
noss 0
root prz
se 0
seed 737548365
[40]:
## a dictionary for storing treemix objects
tdict = {}
## iterate over values of m
for rep in xrange(4):
for mig in xrange(4):
## create new treemix object copy
name = "mig-{}-rep-{}".format(mig, rep)
tmp = tmix.copy(name)
## set params on new object
tmp.params.m = mig
## run treemix analysis
tmp.run()
## store the treemix object
tdict[name] = tmp
[43]:
import toyplot
## select a single result
tmp = tdict["mig-1-rep-1"]
## draw the tree similar to the Treemix plotting R code
## this code is rather new and will be expanded in the future.
canvas = toyplot.Canvas(width=350, height=350)
axes = canvas.cartesian(padding=25, margin=75)
axes = tmp.draw(axes)
[44]:
import toyplot
import numpy as np
## plot many results
canvas = toyplot.Canvas(width=800, height=1200)
idx = 0
for mig in range(4):
for rep in range(4):
tmp = tdict["mig-{}-rep-{}".format(mig, rep)]
ax = canvas.cartesian(grid=(4, 4, idx), padding=25, margin=(25, 50, 100, 25))
ax = tmp.draw(ax)
idx += 1
ABBA-BABA admixture inference
[47]:
bb = ipa.baba(
data=min4.outfiles.loci,
newick="analysis-raxml/RAxML_bestTree.min13",
)
[55]:
## check params
bb.params
[55]:
database None
mincov 1
nboots 1000
quiet False
[53]:
## generate all tests from the tree where 32082 is p4
bb.generate_tests_from_tree(
constraint_dict={
"p4": ["32082_przewalskii"],
"p3": ["30556_thamno"],
}
)
36 tests generated from tree
[54]:
## run the tests in parallel
bb.run(ipyclient=ipyclient)
[####################] 100% calculating D-stats | 0:00:37 |
[59]:
bb.results_table.sort_values(by="Z", ascending=False).head()
[59]:
| dstat | bootmean | bootstd | Z | ABBA | BABA | nloci | |
|---|---|---|---|---|---|---|---|
| 12 | 0.198 | 0.199 | 0.027 | 7.291 | 593.156 | 396.844 | 10629 |
| 27 | -0.208 | -0.207 | 0.031 | 6.648 | 370.500 | 565.000 | 10365 |
| 20 | 0.208 | 0.208 | 0.031 | 6.625 | 565.000 | 370.500 | 10365 |
| 16 | 0.186 | 0.186 | 0.032 | 5.830 | 555.250 | 381.250 | 10243 |
| 26 | -0.186 | -0.186 | 0.033 | 5.708 | 381.250 | 555.250 | 10243 |
[63]:
## most significant result (more ABBA than BABA)
bb.tests[12]
[63]:
{'p1': ['35236_rex'],
'p2': ['35855_rex', '40578_rex'],
'p3': ['30556_thamno'],
'p4': ['32082_przewalskii']}
[64]:
## the next most signif (more BABA than ABBA)
bb.tests[27]
[64]:
{'p1': ['40578_rex'],
'p2': ['35236_rex'],
'p3': ['30556_thamno'],
'p4': ['32082_przewalskii']}
BPP – species tree inference/delim
[68]:
## a dictionary mapping sample names to 'species' names
imap = {
"prz": ["32082_przewalskii", "33588_przewalskii"],
"cys": ["41478_cyathophylloides", "41954_cyathophylloides"],
"cya": ["30686_cyathophylla"],
"sup": ["29154_superba"],
"cup": ["33413_thamno"],
"tha": ["30556_thamno"],
"rck": ["35236_rex"],
"rex": ["35855_rex", "40578_rex"],
"lip": ["39618_rex", "38362_rex"],
}
## optional: loci will be filtered if they do not have data for at
## least N samples/individuals in each species.
minmap = {
"prz": 2,
"cys": 2,
"cya": 1,
"sup": 1,
"cup": 1,
"tha": 1,
"rck": 1,
"rex": 2,
"lip": 2,
}
## a tree hypothesis (guidetree) (here based on tetrad results)
## for the 'species' we've collapsed samples into.
newick = "((((((rex, lip), rck), tha), cup), (cys, (cya, sup))), prz);"
[69]:
## initiata a bpp object
b = ipa.bpp(
name=min4.name,
data=min4.outfiles.alleles,
imap=imap,
minmap=minmap,
guidetree=newick,
)
[70]:
## set some optional params, leaving others at their defaults
## you should definitely run these longer for real analyses
b.params.burnin = 1000
b.params.nsample = 2000
b.params.sampfreq = 20
## print params
b.params
[70]:
burnin 1000
cleandata 0
delimit_alg (0, 5)
finetune (0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01)
infer_delimit 0
infer_sptree 0
nsample 2000
sampfreq 20
seed 12345
tauprior (2, 2000, 1)
thetaprior (2, 2000)
usedata 1
[71]:
## set some optional filters leaving others at their defaults
b.filters.maxloci=100
b.filters.minsnps=4
## print filters
b.filters
[71]:
maxloci 100
minmap {'cys': 4, 'rex': 4, 'cup': 2, 'rck': 2, 'cya': 2, 'lip': 4, 'sup': 2, 'tha': 2, 'prz': 4}
minsnps 4
run BPP
You can either call ‘write_bpp_files()’ to write input files for this data set to be run in BPP, and then call BPP on those files, or you can use the ‘.run()’ command to run the data files directly, and in parallel on the cluster. If you specify multiple reps then a different random sample of loci will be selected, and different random seeds applied to each replicate.
[75]:
b.write_bpp_files()
input files created for job min4 (100 loci)
[76]:
b.run()
submitted 2 bpp jobs [min4] (100 loci)
[77]:
## wait for all ipyclient jobs to finish
ipyclient.wait()
[77]:
True
[90]:
## check results
## parse the mcmc table with pandas library
import pandas as pd
btable = pd.read_csv(b.files.mcmcfiles[0], sep="\t", index_col=0)
btable.describe().T
[90]:
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| theta_1cup | 2000.0 | 0.002 | 4.720e-04 | 1.110e-03 | 1.949e-03 | 0.002 | 0.003 | 0.005 |
| theta_2cya | 2000.0 | 0.001 | 4.587e-04 | 1.944e-04 | 9.056e-04 | 0.001 | 0.002 | 0.003 |
| theta_3cys | 2000.0 | 0.001 | 2.569e-04 | 7.718e-04 | 1.091e-03 | 0.001 | 0.001 | 0.002 |
| theta_4lip | 2000.0 | 0.002 | 3.075e-04 | 9.518e-04 | 1.619e-03 | 0.002 | 0.002 | 0.003 |
| theta_5prz | 2000.0 | 0.007 | 7.235e-04 | 4.556e-03 | 6.150e-03 | 0.007 | 0.007 | 0.009 |
| theta_6rck | 2000.0 | 0.002 | 4.692e-04 | 1.090e-03 | 1.860e-03 | 0.002 | 0.002 | 0.004 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| tau_13rexliprcktha | 2000.0 | 0.002 | 2.376e-04 | 1.470e-03 | 1.854e-03 | 0.002 | 0.002 | 0.003 |
| tau_14rexliprck | 2000.0 | 0.002 | 2.367e-04 | 1.191e-03 | 1.781e-03 | 0.002 | 0.002 | 0.003 |
| tau_15rexlip | 2000.0 | 0.002 | 2.466e-04 | 9.497e-04 | 1.717e-03 | 0.002 | 0.002 | 0.003 |
| tau_16cyscyasup | 2000.0 | 0.005 | 4.718e-04 | 3.503e-03 | 4.881e-03 | 0.005 | 0.006 | 0.007 |
| tau_17cyasup | 2000.0 | 0.002 | 7.494e-04 | 3.230e-04 | 1.270e-03 | 0.002 | 0.002 | 0.005 |
| lnL | 2000.0 | -13806.667 | 2.362e+01 | -1.389e+04 | -1.382e+04 | -13806.152 | -13790.543 | -13733.031 |
26 rows × 8 columns
Success – you’re finished
Now that you have an idea of the myriad ways you can assembly your data, and some of the downstream analysis tools you are ready to explore RAD-seq data.